This is an old revision of the document!
Table of Contents
Pathway Prediction in Batch Mode
Pathway prediction in enviPath can now be performed in batch mode. This feature accepts a .tsv file containing SMILES representations and compound names as input. It processes the file and returns a .tsv output containing the predicted transformation products (TPs) along with relevant metadata. Batch mode is designed to support experimentalists in generation of suspect screening lists, ideal for non-target analysis.
Input requirements
The batch prediction tool requires the following three inputs:
- Input file: A .tsv file containing SMILES strings and compound names, without a header. A template input file can be downloaded here. There is an upper limit of 250 compounds per file. If your input file contains more than 250 compounds, please split it into smaller batches before running the predictions.
- Relative Reasoning model: The relative reasoning model that will be used for the predictions. A default model is pre-selected, but users can also choose a different model if preferred.
- Number of transformation products (TPs): The maximum number of TPs to predict per input compound. The default value is set to 30 TPs per compound, but users can choose any number between 1 and 50.
Algorithm Description
The batch mode uses a greedy-search algorithm where:
- Compounds are represented as Nodes
- Biotransformation reactions are represented as edges, each assigned a weight corresponding to the predicted probability of the reaction occurring, based on available data and competing reaction pathways.
The probability of a reaction (p_edge) is obtained from the machine learning-based relative reasoning algorithm.
The probability of a child node generated during pathway search, also known as the combined probability, is determined based on the probability of the parent node and the reaction probability, as illustrated in the below figure.
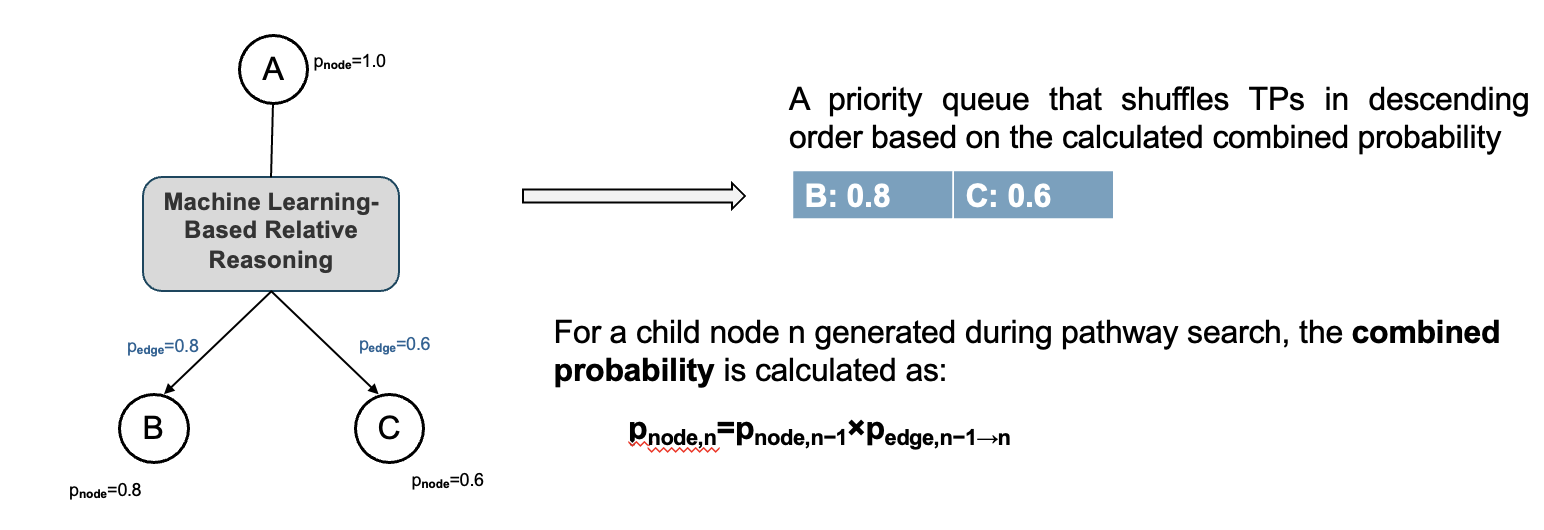
All transformation products (TPs) resulting from reactions with a probability greater than zero are stored in a priority queue, ordered in descending order based on their combined probability. The greedy algorithm makes a locally optimal decision at each step by always expanding the node with the highest combined probability first.
Pathway exploration continues until either the user-defined threshold for the number of TPs is reached, or there are no more TPs remaining in the priority queue to expand.
In some cases, compounds may have fewer predicted TPs than the user-defined threshold, or no TPs predicted at all. This can happen for either of the following reasons:
- No applicable transformation rules were available beyond a certain point, causing the pathway search to stop.
- The probabilities for all predicted transformation products were zero, preventing further expansion.
