This is an old revision of the document!
Table of Contents
Pathway Prediction
Applicable Domain
For all compounds we calculate an applicability score that indicates whether the compound is similar to the training set, in terms of fingerprints and compatible rules. Additionally, all functional groups containing hetero atoms are highlighted in green (red), indicating that they are (not) sufficiently represented in the training set. The evaluation of the applicability domain criteria can be visualized in the pathway view (see example).
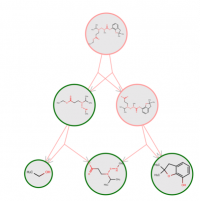
Default Prediction
The SMILES representation of a compound can serve as the input in the dialog on the home page of enviPath. After clicking “Go!”, if the pathway of your compound has already been stored in enviPath, you will have options to either view existing pathways or predicting new pathways. Otherwise, new pathways will be predicted. Prediction performed in this way will use the default setting (i.e., mode and truncation strategy) in enviPath.

